Cells release different types of small-sized membrane vesicles (from 30nm to 1000nm) as a way of communicating with other cells and remove unwanted cell contents; these vesicles are collectively known as extracellular vesicles (EVs). Here we share how we have optimized the imaging of purified EVs to study their size, content and behavior using super-resolution microscopy.
What can we define as extracellular vesicles (EVs)?
Extracellular vesicles have been classically further sub-classified according to their size and origin, as exosomes or microvesicles, among others. However, after the recent International Society for Extracellular Vesicles (ISEV) annual conference in Kyoto, the society issued a statement endorsing “the use of the generic term extracellular vesicle (EV) for particles naturally released from the cell that are delimited by a lipid bilayer and that cannot replicate (do not contain a functional nucleus).” Importantly, they highlighted that, unless specific markers can be used to reliably establish the subcellular origin of different EVs or these are imaged live while being released from a cell, scientists should stay clear of using terms such as exosomes or microvesicles – as these can present contradictory definitions and inaccurate biogenesis associations. According to their guidelines, EV subtypes should be described with the following criteria:
⇒ The physical characteristics of EVs, such as size: small EVs (sEVs) <200nm or medium/large EVs (m/lEVs)
⇒ The biochemical composition: tetraspanins CD63+/CD81+- EVs, Annexin A5-stained EVs, etc.
⇒ The descriptions of conditions or cell of origin: podocyte EVs, hypoxic EVs, large oncosomes, apoptotic bodies, multivesicular bodies.
Read more about the nomenclature of EVs in the statement published by the ISEV here.
Why are scientists looking at extracellular vesicles?
Extracellular vesicles (EVs) can be found in serum, extracellular media and, in all biological fluids, including cerebrospinal fluid, mucus, milk or urine. The comparison between cells grown in regular medium or with so-called ‘exosome-depleted’ serum made apparent the ability of EVs to interfere with biological functions (e.g. in cell proliferation) (Kornilov et al. 2018). This has led to a more exhaustive exploration of EV function, with a field that is rapidly growing.
EVs are highly stable membrane-enclosed particles that can transport different molecules, such as RNA, DNA or proteins; these molecules can either be membrane-bound or enclosed within the vesicle. EVs can also have a variety of characteristics according to the cell type releasing them, which can have a significant impact on their composition. Circulating EVs are essentially now understood as small long-distance signaling units travelling around our body. Therefore, scientists are interested in better characterizing EVs and exploring their signaling functions in a range of diseases, such as cancer (metastasis) or neurosciences, and in crucial immune responses to physiological and pathological processes.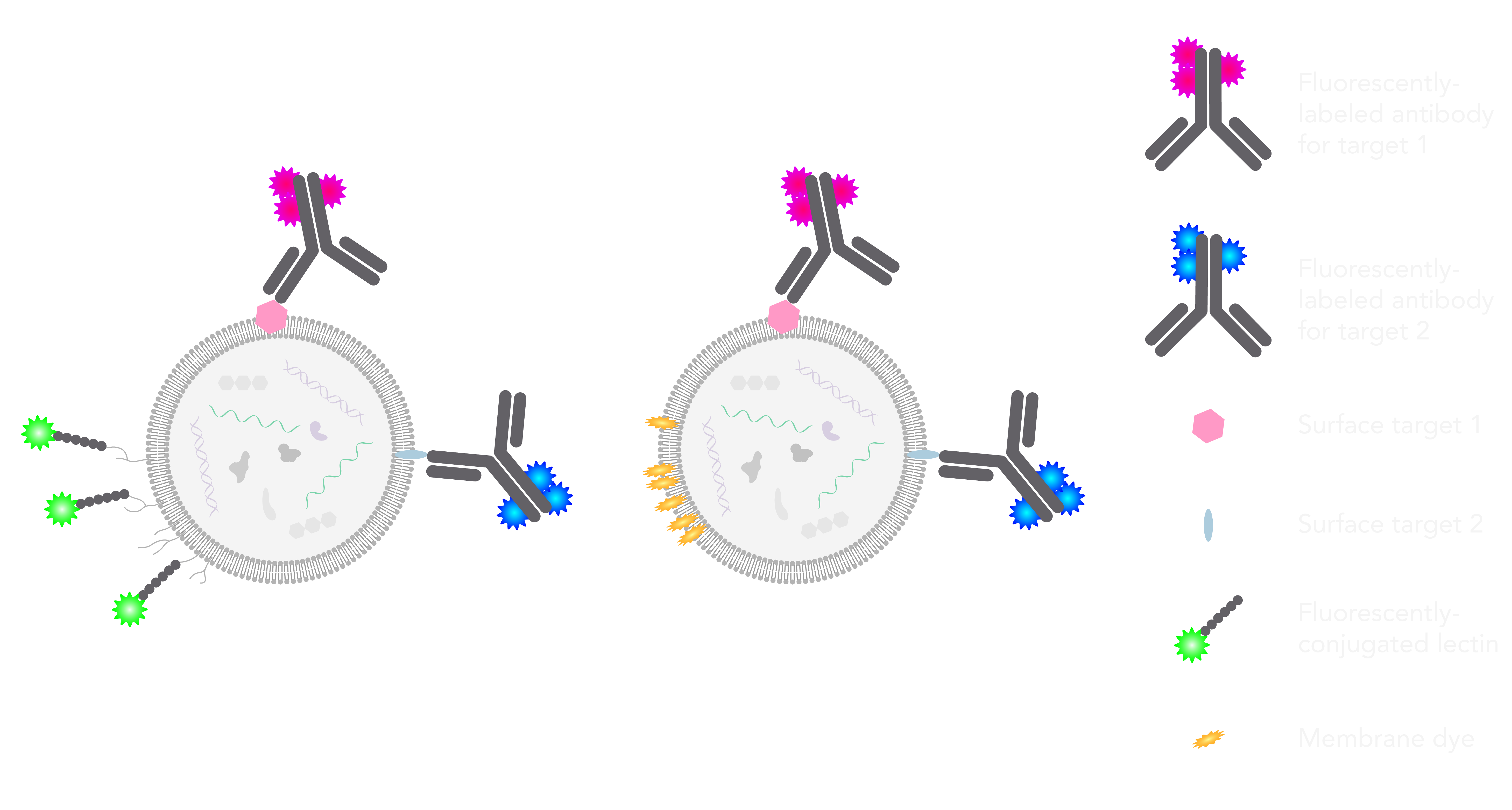
Furthermore, scientists are exploring the use of artificially-created EVs to transport and deliver specific bits of RNA or DNA to certain tissues or cell types. This could help get certain drugs to cross the blood-brain barrier, for neuronal disorders, and potentially make a big difference in bio-availability and treatment efficiency.
What are the current challenges and limitations when studying and visualizing EVs?
One of the main challenges of working with EVs is purifying them without damaging their morphology and composition. Several experts in the field have shared their knowledge on the best procedures for this step and, although work is still required to improve this, researchers are focusing on ultracentrifugation and size-exclusion based isolation methods.
Until recently, some of the main tools available to characterize EVs after purification and obtain significant information on their size, composition and sub-populations have been the ones described in brief below. These techniques have different limitations and studying extracellular vesicles at high resolution, while obtaining relevant functional and structural information remains a challenge.
- Proteomic analysis: used to characterize the composition of EVs by looking at certain defined or novel protein markers of different EV populations. These include Western blot and mass spectrometry. Main limitations: high concentrations of starting material and antibodies required, heavily affected by non-specific antibody binding. Not possible to obtain dynamic data, sizing nor morphological information.
- Electron Microscopy (EM): EM has been the standard imaging technique used to visualize nanoscale details of EVs until recent advances in light microscopy. Main limitations: EM sample preparation can be quite harsh on EVs, as it is likely to affect their morphology (there are reports of oval, round and even cup-shaped EVs), consequently having an impact on the consistency of the results obtained. Additionally, EM relies on specialized knowledge and reagents, and is a multi-step procedure that can be time consuming and, therefore, could prove difficult when carrying out multiplexed imaging.
- Imaging and nano-flow cytometry: a flow-based analysis of particles passing through the optical field has been used to study EV populations. Main limitations: low spatial-resolution as most flow cytometers are not designed to detect vesicles at the lower end of the EV size range (with a few exceptions). In addition, these techniques heavily rely on the specificity of antibodies, without the possibility of assessing the spatial distribution of fluorescence signal, which can potentially result in data misinterpretation due to false positives or unspecific binding of EVs to the beads.
In recent years, additional efforts have been put towards methods for capturing EVs in solution, using plate-based immunocapture techniques that rely on antibodies to bind extracellular vesicles proteins. The external composition of the EV surface layer, however, can make it difficult to assess specificity and obtain consistent data.
Our approach to imaging purified extracellular vesicles
To date, super-resolution fluorescence imaging of EVs has not been a tool available to scientists in the field. This is important not only for having the ability to visualize single molecules, but also to be able to analyze different EV populations, which cannot be done with conventional microscopy techniques. Super-resolution microscopy is now enabling scientists to assign the localization of various EV markers as being either present on the surface or within the vesicle as cargo.
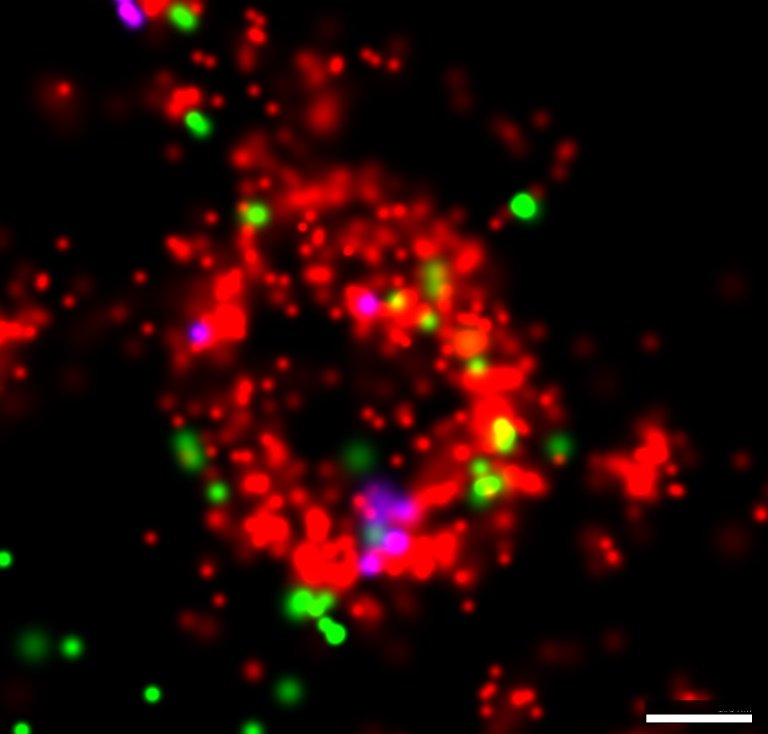
Three-color extracellular vesicles isolated from keratinocyte culture media labeled with lectin stain (EV membrane in red) and fluorescently conjugated antibodies against tetraspanins (CD63 in green and CD81 in magenta) imaged with the Nanoimager using dSTORM. Scale bar is 100nm.
In the ONI lab, the imaging team wanted to first find a way to label the EV membrane. We tried a number of dyes to select those suitable for dSTORM (lipophilic ones have the tendency to precipitate and generate artefacts, and/or aren’t chemically suitable for dSTORM). We decided to narrow it down to a surface glycoprotein-binding lectin coupled to Alexa Fluor 647 to label the sugars groups on the EV surface (as pictured in the top schematic, green label). Another suitable membrane label that we identified is a class of lipophilic dyes commercially known as CellMask™ dyes, which accumulate at the membrane bilayer and have chemical structures suitable for super-resolution imaging (top schematic, yellow label).
Even after thorough optimization steps (see our blog post of 9 tips to optimize your immunofluorescence staining protocol), one of the remaining challenges was to reduce the non-specific background staining bound to the coverslip. To this end, we used an in-house protocol to label EVs in solution. We then plated the EVs onto BSA-treated chamber coverslips, which minimizes the binding of free labels. EVs were finally left to settle and attach to the coverslip, a step that provides the stability essential for high-precision localization of molecules. After washing gently and adding the appropriate dSTORM buffers, the EVs were imaged at super-resolution with the Nanoimager.
The Nanoimager offers the possibility to combine different light-microscopy based techniques to study EVs, including dSTORM imaging with a resolution of around 20nm. One can get a general overview of a sample containing many EVs, colocalization data, counting, sizing and tracking of EVs – it’s a compact solution for an exhaustive cost-effective study of EVs! While dSTORM allows the visualization of details at the nanoscale level, important multi-parametric population data can be inferred from the free diffusion of EVs with two fluorescent labels. This will allow the classification of vesicles according to size and presence or absence of markers.
Lessons learnt
In our case, EVs were initially purified from cell culture media using commercially available kits (precipitation-based, for instance) for extracellular vesicle purification. As mentioned, purification is a tricky step that needs to be optimized to ensure that the shape and composition of the vesicles remains as close to physiological possible.
We have recently optimized the protocol for 3-color imaging to visualize a wide population EVs and quantify sub-populations using two separate markers. As we found out, EV populations can display heterogeneity at multiple levels, making a general labeling strategy difficult to attain. Based on our observations we would recommend being cautious when choosing your labeling strategy, particularly when comparing EVs of different origin. Here are a couple of general tips to bear in mind when planning your EV experiments:
Tip #1: When using antibodies, beware of specificity of the antibodies. Some of the components on the surface around EVs are prone to bind proteins in a non-specific manner.
Tip #2: Keep imaging times as short as possible (imaging happens quite shortly after isolation). Remember that EVs are not fixed, which makes them less stable but more intact and less artefact-prone.
Regarding the biology of EV, there are many challenges and unknown mechanisms, such as those related to entry into the cell (membrane fusion or endocytosis) and dynamic interactions in living cell, especially when considering different cell types. Future studies on the profiling of extracellular vesicles using mass-spectrometry or sequencing will also help widen a field that is opening up with some really exciting discoveries!
Download our EV guide below on how to visualize and study extracellular vesicles with super-resolution imaging using the Nanoimager or get in touch using the form at the bottom of the page.
Share this article:
-1.png?width=717&name=image%20(4)-1.png)