What is super-resolution microscopy?
The ability to resolve small structures is limited in fluorescence microscopy by the properties of light and the inability of optical systems to focus waves to single points below a certain diameter. Thus, optical resolution, or the ability to separate two points that are close together, is limited by the diffraction limit, which makes objectives smaller than 200 nm look “blurry.”
In the early 90s, lateral resolution was improved by confocal microscopy, which uses optical sectioning through out-of-focus light rejection. It was only in the early 2000s when the so-called diffraction barrier was beaten with new super-resolution microscopy technologies that used newly developed dyes, the use of light, and new imaging concepts as well as advanced computational tools. Their recognition culminated with the 2014 Nobel Prize in Chemistry award for some of the foundational work of such super-resolution microscopy methods.
Super-resolution microscopy techniques have been traditionally classified into how illumination is controlled, with the three main techniques being: 1. SMLM (single-molecule localization microscopy) techniques, 2. STED and 3. SIM. More recent developments have also combined SMLM with illumination principles from other techniques, these include 4Pi single-molecule switching (4Pi-SMS), MINFLUX and SIMFLUX, which further increase localization precision and, thus, lateral resolution. Other techniques that can help facilitate improvements in resolution, such as expansion microscopy, or those called ‘soft’ super-resolution techniques, like photon reassignment, Airy scan or iSIM, are also discussed within this group. Each method has its strengths and weaknesses; these have been widely reviewed by experts in the field 1-3.
SMLM (Single-molecule localization microscopy)
SMLM is the umbrella term given to various techniques that use the photochemical properties of certain fluorophores to switch them between a dark and an emissive state. This allows a small subset of labeled molecules to be detected at any given time, enabling the signal isolation of each molecule within a diffraction-limited region and resulting in the localization of each fluorescent molecule at the center of a Gaussian fitting of the point spread function (PSF) formed due to diffraction of light. SMLM retains the advantage linked to conventional fluorescence microscopy while circumventing the diffraction limit, allowing researchers to visualize biological structures, subcomplexes and proteins with a resolution greater than 20 nm, with ten times better detail than with conventional fluorescence techniques.
Figure 1. Animation of how SMLM works. Initial frames show a diagram of a biological structure and how conventional microscopy would resolve this as a blurry ring. On the other hand, with SMLM fluorophores quickly go from an off to an on state, almost one by one, allowing to position their localization through a Gaussian fit of the PSF, thus, resolving the structure with high resolution.
SMLM techniques include:
- Stochastic Optical Recontruction Microscopy (STORM, also known as dSTORM): Relies on the stochastic activation of single fluorophores that blink from “off” to “on” and quickly back to “off”. The process is repeated many times, activating just one molecule within a diffraction-limited region at any given time.
Best for: very high resolution, molecular distributions in fixed cells
Requires: tagging with compatible organic dyes, high brightness and blinking buffer.
- Point Accumulation for Imaging in Nanoscale Topology (PAINT): Single-molecule localizations achieved using transient binding of fluorophores to targeted proteins. PAINT is commonly performed with DNA strands < 10 nt. Like other SMLM techniques, it can reach spatial resolutions of 20 nm.
Best for: Biomolecular organization and quantification, in fixed samples.
Requires: Specific transient binding molecules linked to fluorescent dyes
- PhotoActivated Light Microscopy (PALM): Uses photoactivatable fluorophores to resolve spatial details of tightly packed molecules. Once activated, fluorophores emit for a short period but eventually bleach. The laser stochastically activates fluorophores until all have emitted.
Best for: Protein dynamics, organization and quantification, in fixed and live samples.
Requires: Genetic encoding using photoswitchable fluorescent proteins, low brightness.
- Single Particle Tracking (SPT): Allows the motion of individual particles to be followed in vitro or in living cells to obtain information on their dynamic behavior over time. Trajectories can be extracted with a resolution of up to milliseconds.
Best for: Protein dynamics and tracking, in live samples or particles in solution.
Requires: Genetic encoding using photoswitchable fluorescent proteins, low brightness, low tagging density.
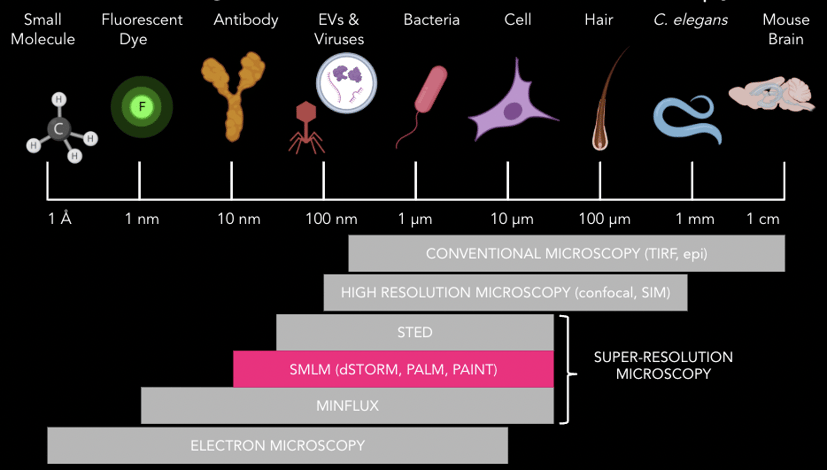
Figure 2. Schematic representation of biological specimens across scales, from centimeters, down to nanometers and beyond. The diffraction limit of light (Abbe’s law) is indicated at around 200 nm. Super-resolution techniques, such as SMLM, are able to resolve structures over ten times smaller than that, compared to conventional fluorescence microscopes.
Super-resolution microscopy vs. electron microscopy
For decades, electron microscopy (EM) has been the only available imaging method for those aiming to visualize small biological structures in the nanometer scale. Despite its ability to achieve better orders of magnitude resolution than fluorescence microscopy, EM remains a complex technique due to its laborious sample preparation, difficulty in labeling and expertise needed to carry out imaging and identification of structures.
Super-resolution microscopy makes imaging at the nanoscale more accessible, with the ability to prepare samples in a similar fashion as in conventional fluorescence microscopy for most techniques. It enables specific labeling of targets and it results in much more quantitative results than electron microscopy, allowing to determine molecule numbers with given techniques, structure size, and track molecules if techniques compatible with live imaging are used. Overall, it means that super-resolution microscopy data can uncover small differences across samples, which could be imperceptible to the eye with standard electron microscopy.
While super-resolution microscopy remains challenging to master for some, overall, it is less costly than electron microscopy for specific use cases, such as clinical diagnosis in histopathology labs, where EM is the most expensive single test, and the use of centralized facilities creates unnecessary bottlenecks. Super-resolution microscopy techniques, such as dSTORM, have the great potential to result in faster diagnosis, increased sensitivity and confidence for some diseases.
More recently, several advances are combining the strengths of different imaging techniques to combine benefits, for instance, the development of correlative electron microscopy (CLEM), such as advanced FIB-SEM, which allow powerful CLEM workflows, or super-resolution imaging under cryo-conditions.
How can super-resolution microscopy benefit my research?
There is a lot of information that one can extract from super-resolution microscopy. The most obvious one is the ability to visualize molecular structures or cellular complexes with great level of detail and obtain high-resolution images. However, the power of super-resolution imaging lies in its ability to obtain high amounts of quantitative information on the molecule or structures being investigated, including morphology, spatial organization, interaction of molecules, or stoichiometry of macromolecular complexes.
Before starting with super-resolution microscopy, a crucial step is to choose the most suitable technique for your biological question. This will depend on a combination of factors, including the sample type, the size of the structure of interest, and any spatial or temporal resolution requirements. There are dozens of existing super-resolution techniques which may appear intimidating at first, but these techniques are generally just a different protocol, with different dyes or labels, and potentially a slightly different acquisition strategy. The beauty of this is that anyone who has run a protocol in the past is usually capable of running any of the below techniques without significant barriers. Don't be afraid to try and test several to see what works best!
Choosing the best super-resolution technique for me
Most super-resolution techniques improve spatial resolution to subdiffraction resolution, however, some trade-offs are expected depending on the requirements for any given experiment and system. For example, speed of imaging or temporal resolution, need for multicolor imaging, sample thickness, sample sensitivity to photodamage, fixed or live samples, and type of molecular dynamic associated with the biological process under study. In the section above, we covered different SMLM techniques and what these are best for, or their specific requirements. For a comprehensive guide to wider groups of super-resolution methods, we recommend reading literature reviews 1-3. The flow diagram below from Valli and colleagues3 is a helpful simplified guide to choosing a super-resolution technique.
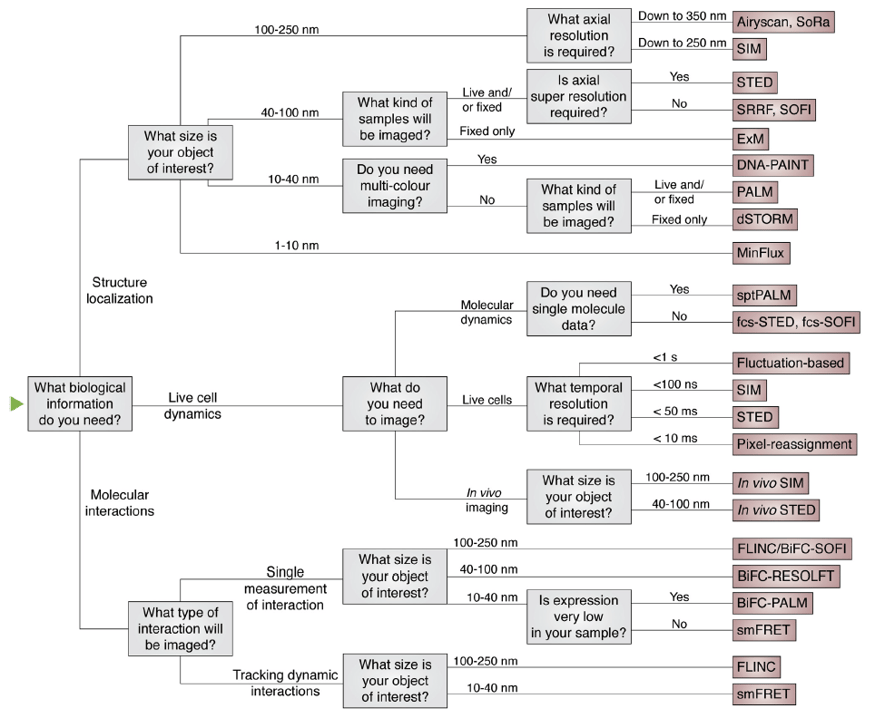
Figure 3. Flow diagram to help choose a super-resolution technique depending on the biological questions in mind and additional imaging requirements. Image from Valli et al. 20203
More recently, some researchers are applying imaging methods consecutively, such as SIM and SMLM on the same widefield imaging platform to obtain information on target localization within a 3D structure. Usually, these are super-resolution experts who are either building their own microscope system or operate within an advanced imaging facility, an option not accessible to most.
There are now several robust commercially available systems for super-resolution microscopy. These have their own strengths and limitations, and it is important to understand the different techniques each system is capable of performing. For instance, ONI’s Nanoimager is a good option when looking at using complementary techniques to investigate a given sample. It combines different imaging techniques within one compact box for examining samples across scales, all the way down to 20 nm. This is unique as most microscopes are dedicated to either super-resolution techniques like dSTORM or SIM imaging, and others to confocal and TIRF imaging. The Nanoimager has incredibly powerful lasers and temperature control, which enable single-molecule localization microscopy in fixed samples (dSTORM, DNA-PAINT), single particle tracking and PALM in live samples, smFRET to look at protein interactions, TIRF or HiLO illumination for signal-to-noise improvement, widefield imaging and confocal imaging for faster imaging. Many of the techniques listed above can be done with the same hardware by only adjusting the reagents, acquisition parameters and analysis workflows. This can allow researchers to interrogate their samples through different means and extract meaningful data from their experiments. Here is a helpful table to help you understand and choose the different techniques available within the Nanoimager.
Extracting meaningful data from super-resolution images
Super-resolution microscopy results in beautiful high-resolution images of biological structures or processes. Highly informative quantitative data can also be extracted from such datasets, including information about single molecule behavior, size, shape, distribution or colocalization, which often enable the detection of changes otherwise imperceptible to the eye. This step is crucial to making the most of super-resolution imaging, but can also be the hardest for some researchers.
Over the last few years, new computational methods to analyze super-resolution microscopy data, including data-driven and machine-learning algorithms are emerging. However, their validation and advancement at the same speed as imaging techniques remain challenging. For SMLM, 2D or 3D point cloud data with millions of localizations is usually generated. These are generally then analyzed using different clustering methods to support quantitation of the image.
Another point to consider is that SMLM data is highly sensitive to noise and can also be biased due to imaging artifacts, non-real or incorrect signal or localizations, which can impact image quality and interpretation. These are usually related to suboptimal sample preparation or acquisition issues, which can relate to 4,5: incomplete or incorrect labeling, detection efficiency, localization uncertainty, background localizations, inaccurate calibration, incorrect filtering of data, blinking artifacts, drift or chromatic aberrations. There are some computational methods to address these issues, and the use of appropriate controls can help minimize these.
If we consider a typical workflow for SMLM imaging, the steps usually are:
- Imaging: a target protein is labeled with a primary and secondary antibody conjugated to a dye or fluorophore (directly-conjugated primary antibodies can be used). Laser light will excite the molecule and produce a fluorescent signal in the form of photons.
- Acquisition: photons from excited molecules are captured by the system’s camera as a signal in the shape of a point spread function (PSF) from diffracted light, which thanks to computational algorithms, results in the localization of a specific molecule in any given frame after Gaussian fitting. Most SMLM methods rely on the temporal separation of emissions of excited fluorophores., where these are rapidly activated at different times, ‘blinks’, ensuring non-overlapping PSFs within a diffraction-limited region. The information of all frames over time and collection of thousands of localization points will gather enough information points throughout the sample.
- Cluster analysis and quantification: different pre-processing steps are performed including drift correction, chromatic aberrations correction, registration, etc. Post-processing tools include clustering algorithms, Voronoi tessellation, nearest neighbor analysis, pair correlation… for more details, we recommend reading literature reviews on the topic. 4,5
There are many popular clustering algorithms, but most approaches rely on some metric of density. In general, localizations that are densely packed form clusters, and these clusters are separated by areas of sparse localizations. In practice, however, this is a complicated problem. For example, due to the varying background across a sample or because some clusters themselves contain smaller clusters.
ONI’s collaborative discovery platform (CODI) offers a few different clustering analyses including DBSCAN, HDBSCAN and our extension of HDBSCAN called Constrained clustering. Behind these methods, we use the concept of a cluster tree, which captures different possible clusters by considering how big clusters may break down into smaller clusters. You can read more about these in a recent blog post Beginners guide to clustering on localization data and watch the recording of a webinar on the same topic by Data Scientist Haraman Johal.
How do I get started with super-resolution imaging?
We are here to help! We have published a follow up article on Getting started with Super-Resolution. We recommend exploring our dSTORM Training Kit to learn the basics of single-molecule localization microscopy, and the dSTORM Discovery Kit to label your favorite proteins in different cell types. You can also browse our website for technical articles, learning resources, and publications that have used our technology on different biological applications.
ONI is on a mission to change this by making super-resolution microscopy more accessible and easy to use. We want to help researchers worldwide access the powerful technology that can allow them to see and understand the world surrounding us with the greatest possible level of detail. Join us in exploring the nanoscale world and pushing scientific boundaries with cutting-edge technology! For any queries and sales support, contact our team at hi@team.oni.
References
- Schermelleh, L., Ferrand, A., Huser, T. et al. Super-resolution microscopy demystified. Nat Cell Biol. 2019; 21: 72–84. https://doi.org/10.1038/s41556-018-0251-8.
- Prakash K, Diederich B, Heintzmann R and Schermelleh L. Super-resolution microscopy: a brief history and new avenues. Phil Trans R Soc A. 2022 Feb; 380: 2220. https://doi.org/10.1098/rsta.2021.0110.
- Valli J, Garcia-Burgos A, Rooney LM, de Melo e Oliveira BV, Duncan RR, Rickman C. Seeing beyond the limit: A guide to choosing the right super-resolution microscopy technique. J Biol Chem. 2021; 297 (1): 100791. https://doi.org/10.1016/j.jbc.2021.100791.
- Khater IM, Nabi IR, Hamarneh G. A Review of Super-Resolution Single-Molecule Localization Microscopy Cluster Analysis and Quantification Methods. Patterns (N Y). 2020 Jun 12; 1(3):100038. doi: 10.1016/j.patter.2020.100038.
- Wu YL, Tschanz A, Krupnik L, Ries J. Quantitative Data Analysis in Single-Molecule Localization Microscopy. Trends Cell Biol. 2020 Nov; 30(11):837-851. doi: 10.1016/j.tcb.2020.07.005.
Share this article:
