Why use Super-resolution Microscopy in Neuroscience?
Each synapse is composed of over 200 proteins and a synaptic cleft, across which communication between cell terminals is mediated. The small size of synapses, under 200 nm in size, and the synaptic cleft, as small as 20-40 nm, can make it difficult to study the molecular mechanisms regulating neurotransmission. That is because their intricate molecular details are ‘invisible’ to conventional, diffraction limited light microscopy techniques.
Super-resolution microscopy is able to break the diffraction limit with techniques such as single-molecule localization microscopy (SMLM), which includes dSTORM imaging, PALM and DNA-PAINT. Multicolor SMLM extends the neuroscience and neurobiology toolbox for characterizing protein organization at the nanometer scale.
Multicolor SMLM extends the neuroscience toolbox for characterizing protein organization at the nanometer scale
Super-resolution microscopy with single-molecule sensitivity enables characterization of the architecture of neuronal synapses, the study of synaptic protein dynamics, molecular interactions, and distribution of neurotransmitters and other biomolecules. Single-molecule imaging is a robust tool to answer the fundamental questions on brain function and dysfunction, and better understand the molecular hallmarks of brain plasticity.
In this example, neuronal synapses were visualized using 3-color dSTORM imaging. Proteins labeled included two presynaptic proteins, Bassoon (synaptic active zone, in magenta) and VGluT1 (synaptic vesicles, in green), and the postsynaptic protein Homer1 (postsynaptic density, in orange). The data revealed the presence and organization of neurotransmitter vesicles in between pre- and postsynaptic protein clusters.
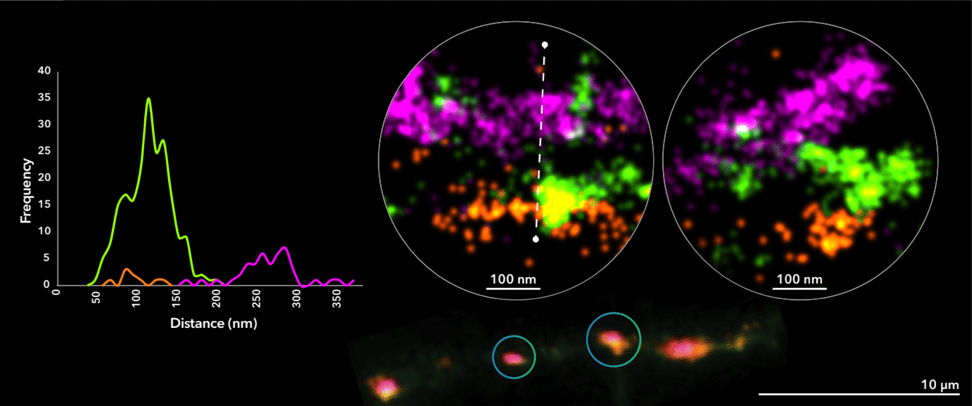
Figure 1. 3-color dSTORM image of neuronal synapses in rat primary cortical neurons cultured for 14 days. Image shows a neuronal process containing several synapses imaged using fluorescently-labeled antibodies against the presynaptic protein Bassoon (magenta), synaptic vesicle marker VGlut1 (green) and the postsynaptic protein Homer1 (orange).
To learn more read the ONI application notes on revealing synapse architecture at a single-molecule level, and quantitative super-resolution imaging reveals nanoscale recruitment of GABAA receptors following Bicuculline treatment.
How should I prepare neuronal cultures for super-resolution imaging?
Culture cell lines for certain brain cell types can be difficult to obtain and maintain in vitro, making it challenging to find truly predictive neural models. Currently, dissociated neurons from brain tissue from rat or mouse embryos (latest days) or from postnatal pups (early days) are the most common sample source for neuronal studies in SMLM. However, these models can be considered artificial environments with low complexity brain architectures. An attractive alternative for neuroscientists is the use of neurons differentiated from iPSCs (induced Pluripotent Stem Cells, generated directly from adult cells), which allows research on human cell lines.
For details to prepare neuronal cultures, you can read our Technical guide for top tips to culture and fix dissociated neurons from mouse or rat brain tissue. In brief:
- Neurons are usually seeded in round #1.5H glass coated coverslips (poly-D-lysine is the most common coating used).
- A density of around 10,000-50,000 cells/cm2 is recommended.
- Submerge coverslips in dishes containing Neurobasal medium supplemented with B-27 Supplement. Alternatively, Ibidi 8-well dishes may be used, depending on the growth conditions of the neuronal culture.
- Neurons are cultured in an incubator on average for 15 days at 37ºC and 5% of CO2.
- After incubation, neurons are fixed using pre-warmed (37ºC) 4% paraformaldehyde (PFA) and 4% glucose in phosphate buffered saline (PBS) for a 5-10 min at room temperature. In some cases, where synapse morphology is important, 0.1% Glutaraldehyde may be included, but this could be at the expense of epitope binding being affected.
How do I stain or label neuronal cultures for imaging?
A usual limitation in neuroimaging is the availability of labeling tools. Available antibodies target the most common and best researched proteins while other players remain unexplored, hindering the detailed analysis of neuronal synapse architecture. Here are a few tips for labeling with or without permeabilization:
- Immunostaining of primary antibodies targeting membrane proteins with extracellular binding sites can be performed before cell permeabilization. Dilute antibodies in blocking buffer, e.g. 1 % of bovine serum albumin (BSA) in PBS, and incubated at room temperature for several minutes (~1h) or overnight at 4ºC.
- To avoid labeling the intracellular pool, surface-expressed protein staining can be performed before fixation. In this case, incubate neurons with antibodies for 10-15 min at 37ºC and fix immediately to avoid protein turnover.
- When labeling intracellular structures, lipid membrane permeabilization is necessary. Use a detergent, such as 0.1-0.5% Triton X-100 in PBS for 5-10 minutes.
- The same buffer used for antibody dilution can be used to block the non-specific binding of staining agents and reduce the background fluorescence of the image. Typically ~1h incubation at room temperature is sufficient.
- The most common method for fluorophore labeling is the use of secondary antibodies conjugated with organic dyes compatible for dSTORM imaging. Immunostaining of multiple targets at once is possible, if different spectral fluorophores are used.
- Primary antibodies directly conjugated to a fluorophore are also available in the market, in which case, a secondary staining is not necessary. Other approaches for labeling include HaloTag® or SNAP-tag® 1, nanobodies, or Fab fragments.
- In neuronal SMLM experiments, it is also possible to use regular tagging methods like in other fluorescence microscopy modalities, such as transfection, viral transduction or electroporation. These methods are recommended for tagging neuronal targets with fluorescent proteins. Photoactivatable Localization Microscopy (PALM) or single-particle tracking (SPT) imaging benefit from this method.
- For PALM imaging, you will need to use photoconvertible fluorescent proteins in order to achieve super-resolution imaging. Read more on fluorophores for PALM imaging.
- More recently, DNA-PAINT or uPAINT have proven to be very suitable for neurological studies. 2, 3 . In this case, oligomers tagged with fluorophores transiently bind to the proteins of interest during imaging .
To learn more on how to mount neuronal cultures depending on the imaging approach used (dSTORM, PAINT, PALM, Single Particle Tracking or SPT) read our Technical guide on Neuronal Culture for Super-Resolution Imaging.
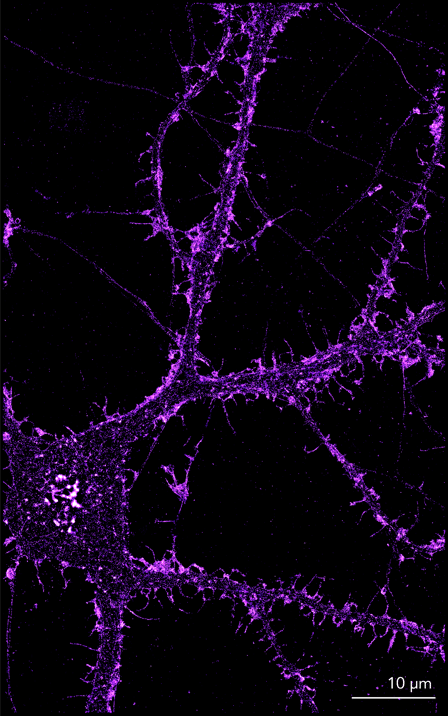
Figure 2. Images show the structure of actin in the dendrites, dendritic spines and axons of primary embryonic rat cortical neurons cultured for 14 days. Samples were pre extracted briefly then fixed using a fixative containing paraformaldehyde and glutaraldehyde. Actin structures were labeled using phalloidin conjugated to AF-647.
How can the Nanoimager help me?
The compact design of the Nanoimager (a smaller footprint than an A4 sheet of paper), its stability (you don’t need an optical table or dark room for it) and overall ease of use, makes it easier for neuroscientists to perform super-resolution imaging anywhere. The Nanoimager is capable of performing up to 4-color imaging, two of them simultaneously, allowing imaging of multiple targets in one acquisition. This feature is very convenient for the study of protein complexes at the neuronal synapse, enables accurate co-localization of proteins in synapses, and paves the way for the study of synapse protein linkage.
Single-molecule localizations acquired during imaging are used to reconstruct a super-resolved image of neuronal protein and synaptic receptor organization 4,5. Clustering algorithms are required to characterize these maps of localizations and extract both structural and quantitative information. CODI, ONI’s cloud-based data analysis platform, can be used to characterize the organization of synaptic proteins at the nanometer scale. Information, such as cluster size, area, or number of localizations per cluster, is extracted.
ONI offers a comprehensive solution for single-molecule localization microscopy in neurons. It is possible to perform different imaging techniques within one same instrument: dSTORM, DNA-PAINT, PALM, and SPT. If you want to learn more about how you can use the Nanoimager for neuroscience research, visit our Neuroscience Hub page and Resources.
To hear first hand from a Nanoimager user, you can watch this webinar from Dr. Harold MacGillavry (Department of Cell Biology at Utrecht University in the Netherlands), where he discusses his work providing new insights into the molecular architecture of the endocytic zone and thereby to reveal novel molecular mechanisms that control local trafficking of glutamate receptors at excitatory synapses.
If you want more information on our dSTORM sample preparation workflow, details on how to prepare coverslips and seed cells, or optimize SMLM imaging and analysis of your neuronal specimens, get in touch with us at hi@oni.team.
References
- Siddig S, Aufmkolk S, Doose, S, Jobin ML, Werner C, Sauer M, Calebiro D. Super-resolution imaging reveals the nanoscale organization of metabotropic glutamate receptors at presynaptic active zones. Sci. Adv. 2020; 6(16).
- Böger C, Hafner AS, Schlichthärle T, Strauss MT, Malkusch S, Endesfelder U, Jungmann R, Schuman EM, Heilemann M. Super-resolution imaging and estimation of protein copy numbers at single synapses with DNA-point accumulation for imaging in nanoscale topography. Neurophotonics. 2019 Jul;6(3):035008.
- Giannone G, Hosy E, Levet F, Constals A, Schulze K, Sobolevsky AI, Rosconi MP, Gouaux E, Tampé R, Choquet D, Cognet L. Dynamic superresolution imaging of endogenous proteins on living cells at ultra-high density. Biophys J. 2010 Aug 9; 99(4): 1303-10.
- Kellermayer B, Ferreira JS, Dupuis J, Levet F, Grillo-Bosch D, Bard L, Linarès-Loyez J, Bouchet D, Choquet D, Rusakov DA, Bon P, Sibarita JB, Cognet L, Sainlos M, Carvalho AL, Groc L. Differential Nanoscale Topography and Functional Role of GluN2-NMDA Receptor Subtypes at Glutamatergic Synapses. Neuron. 2018 Oct 10;100(1):106-119.e7.
- Catsburg LA, Westra M, van Schaik AM, MacGillavry HD. Dynamics and nanoscale organization of the postsynaptic endocytic zone at excitatory synapses. eLife. 2022; 11 (e74387).
Share this article: