Challenge: Revealing dynamic information about viral particles during host-pathogen interactions
 Determining host-pathogen interactions is key to accelerating the development of novel therapeutics. On the aftermath of the last Ebola epidemic, the World Health Organization developed a special tool for “determining which diseases and pathogens to prioritize for research and development in public health emergency contexts”. This tool aims to identify diseases that pose a public health risk because of their epidemic potential and for which, right now, there are no or insufficient countermeasures. From that list, all the pathogenic agents are viruses.
Determining host-pathogen interactions is key to accelerating the development of novel therapeutics. On the aftermath of the last Ebola epidemic, the World Health Organization developed a special tool for “determining which diseases and pathogens to prioritize for research and development in public health emergency contexts”. This tool aims to identify diseases that pose a public health risk because of their epidemic potential and for which, right now, there are no or insufficient countermeasures. From that list, all the pathogenic agents are viruses.
Although vaccines and antiviral compounds have made leaps forward in the treatment of viral pathogens, advancements in diagnostics and therapeutics are still much needed. Helping researchers to characterize dynamic properties of virus particles will enable breakthroughs in managing viral pathogens and in finding new targets to treat viral infection. Key areas could include viral endo/exocytosis, migration towards interaction sites within host cells and locations of virus uncoating.
Solution with the Nanoimager: Locate, count and follow host-pathogen interactions
 Single-particle tracking in live cells allows us to measure the motion and changes in the behavior of fluorescently-labeled viral particles. This means tracking of viral particles as they enter the cell and organelles such as the nucleus is possible. Alternatively, viral egress by lysis, exocytosis or budding through the plasma membrane can also be tracked.
Single-particle tracking in live cells allows us to measure the motion and changes in the behavior of fluorescently-labeled viral particles. This means tracking of viral particles as they enter the cell and organelles such as the nucleus is possible. Alternatively, viral egress by lysis, exocytosis or budding through the plasma membrane can also be tracked.
The Nanoimager enables researchers to learn more about host-pathogen interactions: where viral particles are assembled, accumulate, transiently bind or move to. It allows simultaneous dual-color imaging to easily follow two fluorophore species at the same time. Additionally, with its compact size, ease of use, and remote-controllability, the Nanoimager is the ideal fit for limited spaces, such as a biosafety cabinets, BSL3 labs, and even high-containment BSL4 labs for work with highly-rated pathogenic viruses.>
To precisely measure the effects of viral infection including cell response to pathogens, growth/environment conditions, mutations in the genome and response to drugs, the dedicated NimOS software offers a range of tools for quantitative data analysis. Information about viral particle size, number and diffusion coefficients can be instantly visualized, plotted and saved as a file compatible with other standard data analysis platforms.
Case Study: Virus diffusion in host-pathogen interaction research
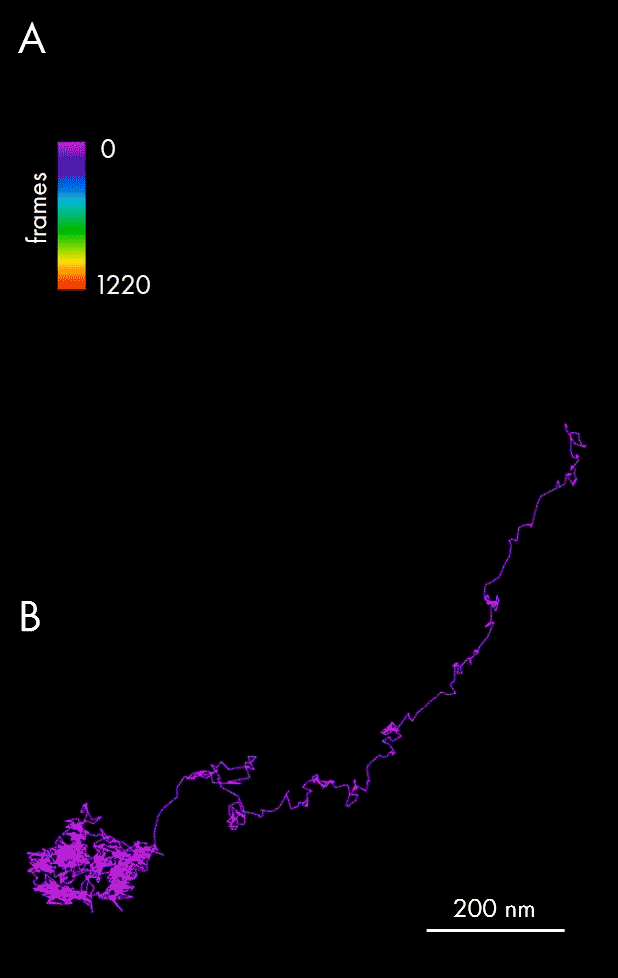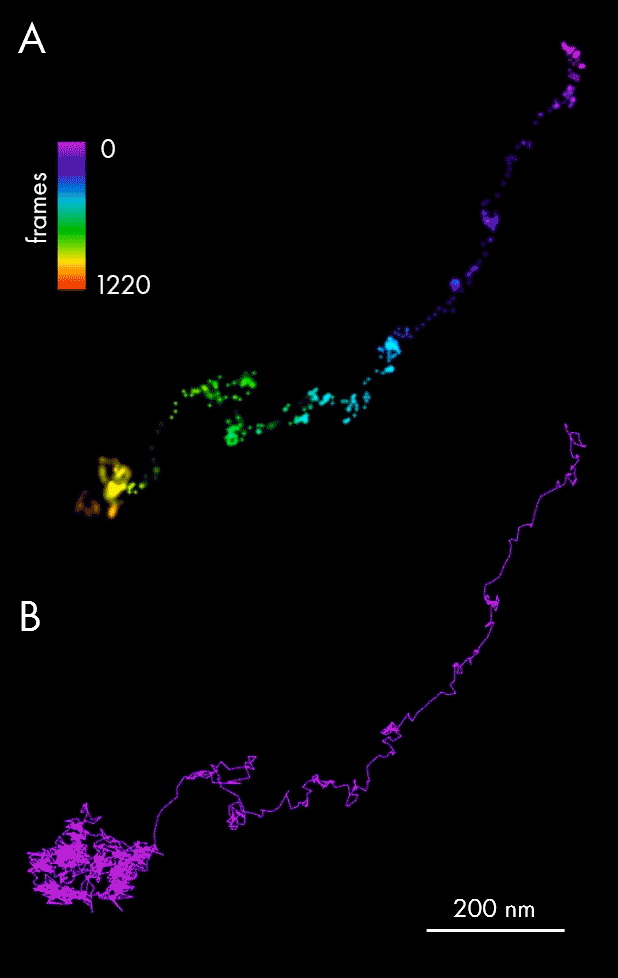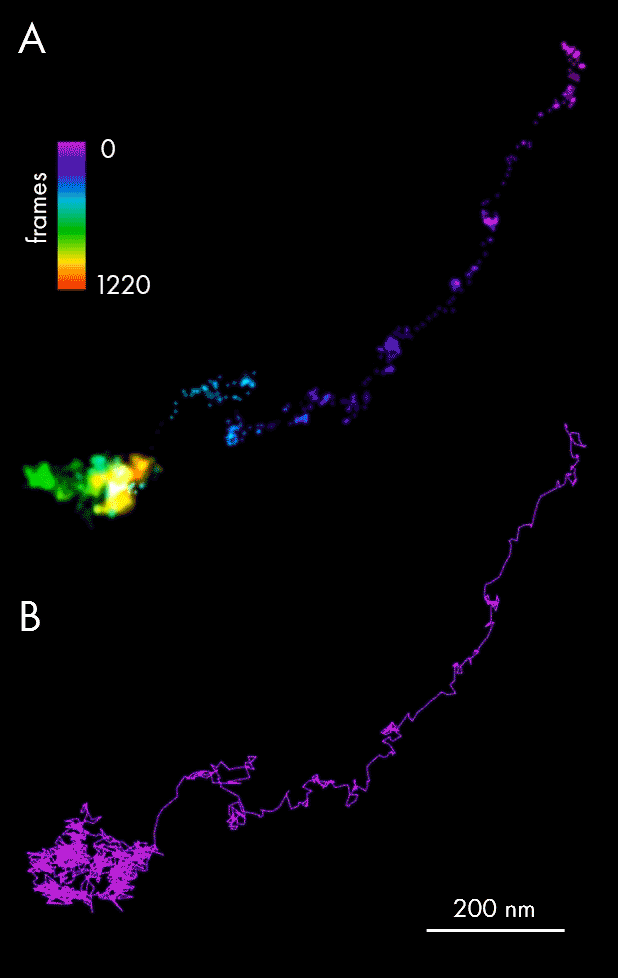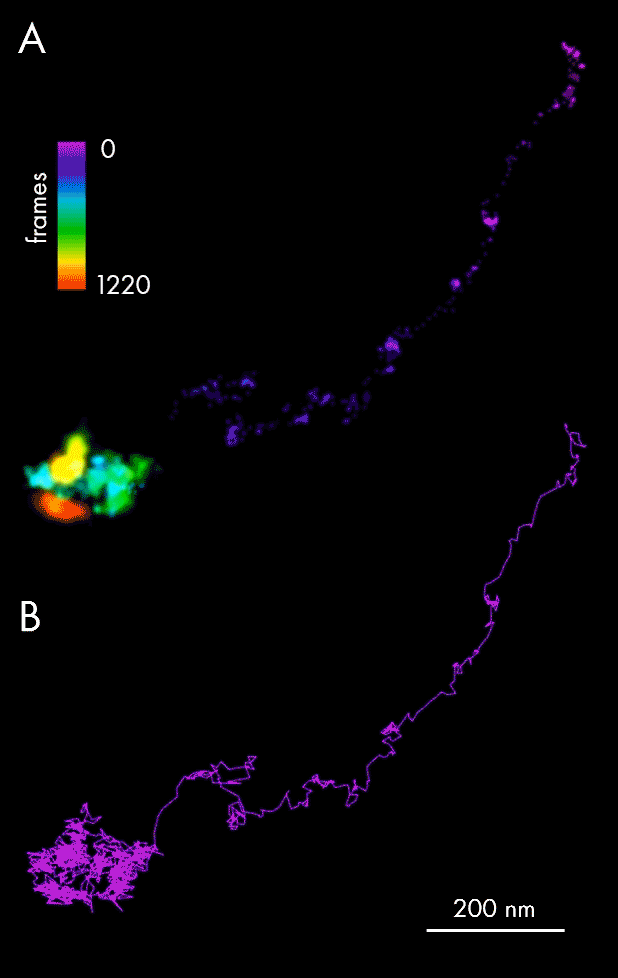
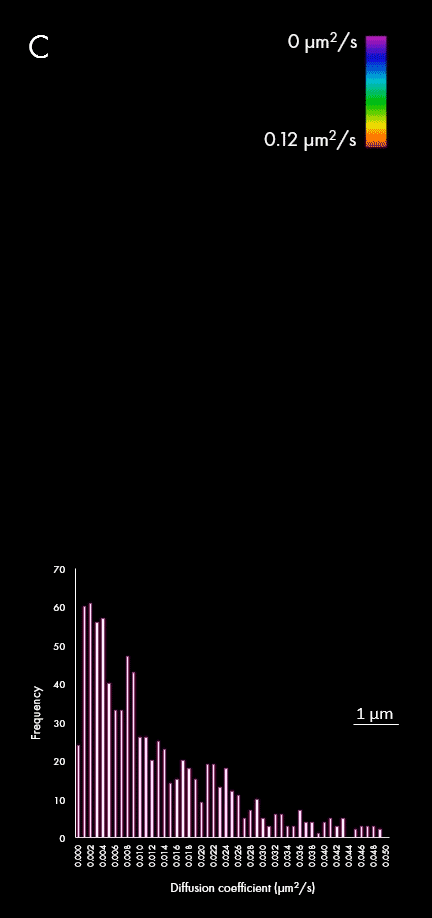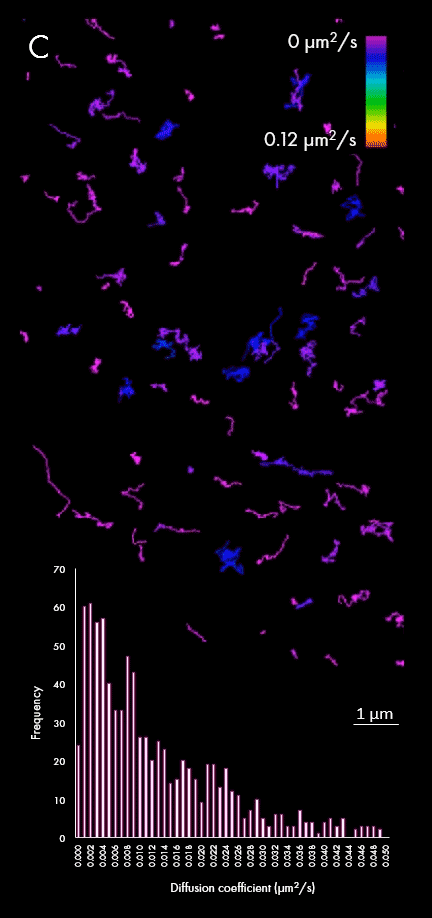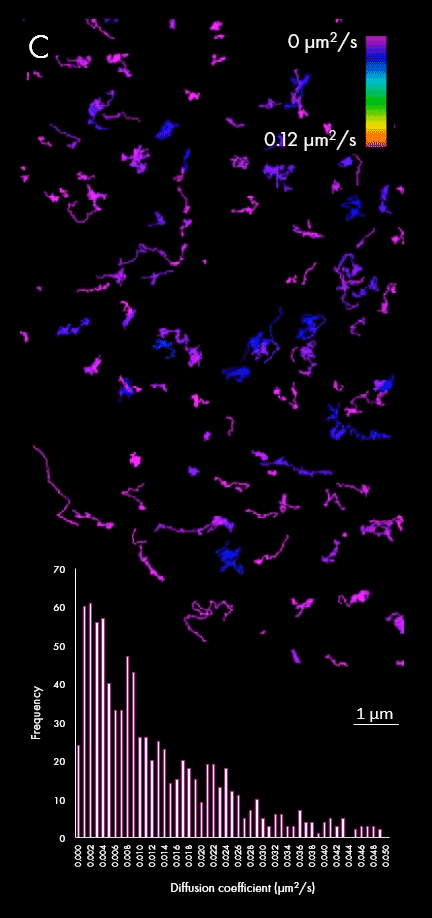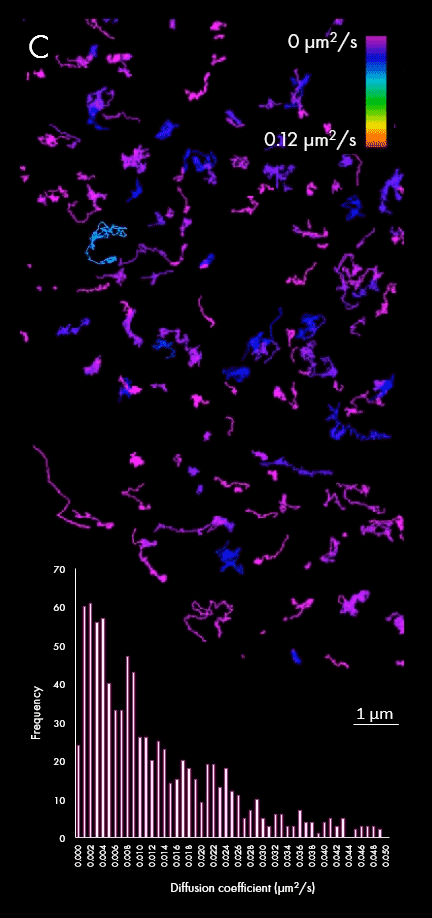 Using single-particle tracking, spatiotemporal host-pathogen interaction dynamics can be defined via localization of viral particles throughout their lifecycle: assembly and maturation in the cell nucleus, envelopment and exit as mature virions. In this example, single-particle tracking analysis allowed tracking of Ebola nucleocapsid-like structures (NCLS-GFP) inside human liver Huh-7 cells.
Using single-particle tracking, spatiotemporal host-pathogen interaction dynamics can be defined via localization of viral particles throughout their lifecycle: assembly and maturation in the cell nucleus, envelopment and exit as mature virions. In this example, single-particle tracking analysis allowed tracking of Ebola nucleocapsid-like structures (NCLS-GFP) inside human liver Huh-7 cells.
Panel A of the figure above presents individual localizations of a viral particle within a cell, appearing over subsequent frames, and color-coded according to the time of appearance. The dedicated NimOS tracking tool was used to plot its trajectory (depicted in panel B, above), allowing the particle’s directionality, speed and position to be followed. The data obtained suggests that the particle was initially moving fast in a clear linear direction, before switching to a random movement within a localized area.
The representation of movement from individual particles allows researchers to determine their directionality, speed and study heterogeneous virus-virus behaviors in different cellular regions or near determined organelles. A large field of view can also capture all the particles within a cell, plotting diffusion coefficients for a wider population, as shown in panel C. This enables researchers to distinguish between subpopulations of viral particles.
The possibility of quickly tracking and quantifying the behavior of thousands of particles, especially when coupled with near real-time analytics and data processing, will provide new insights into these previously poorly characterized particles and speed up the process of drug and vaccine discovery.
Acknowledgment: We would like to thank Professor Stephan Becker, Dr Olga Dolnik, Dr Katharina Grikscheit and their team from Marburg University for sample preparation and insightful discussions.
Read additional case studies about the HBV1 particle tracking and imaging viral particles with dSTORM.
Share this article: